OVERVIEW
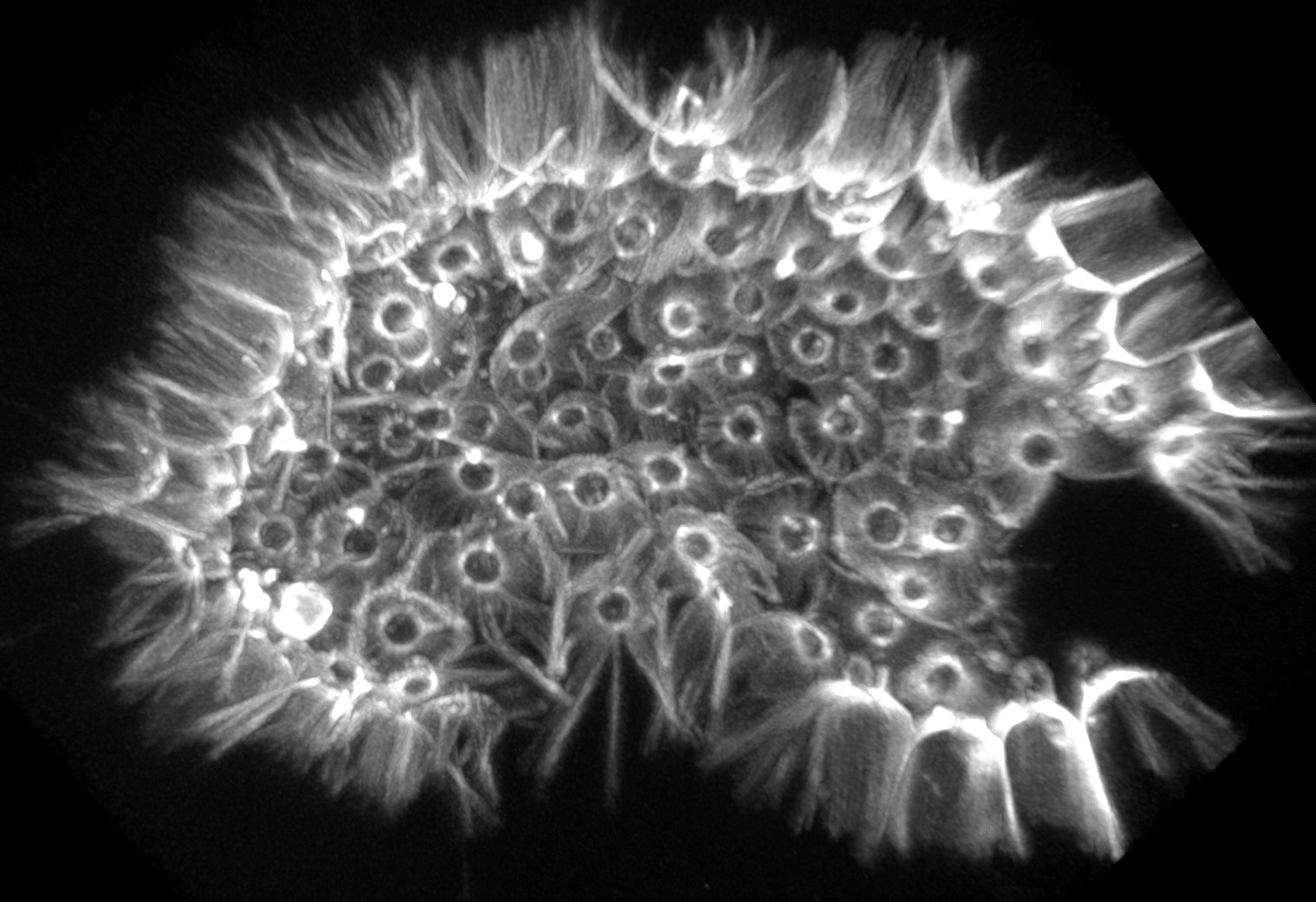
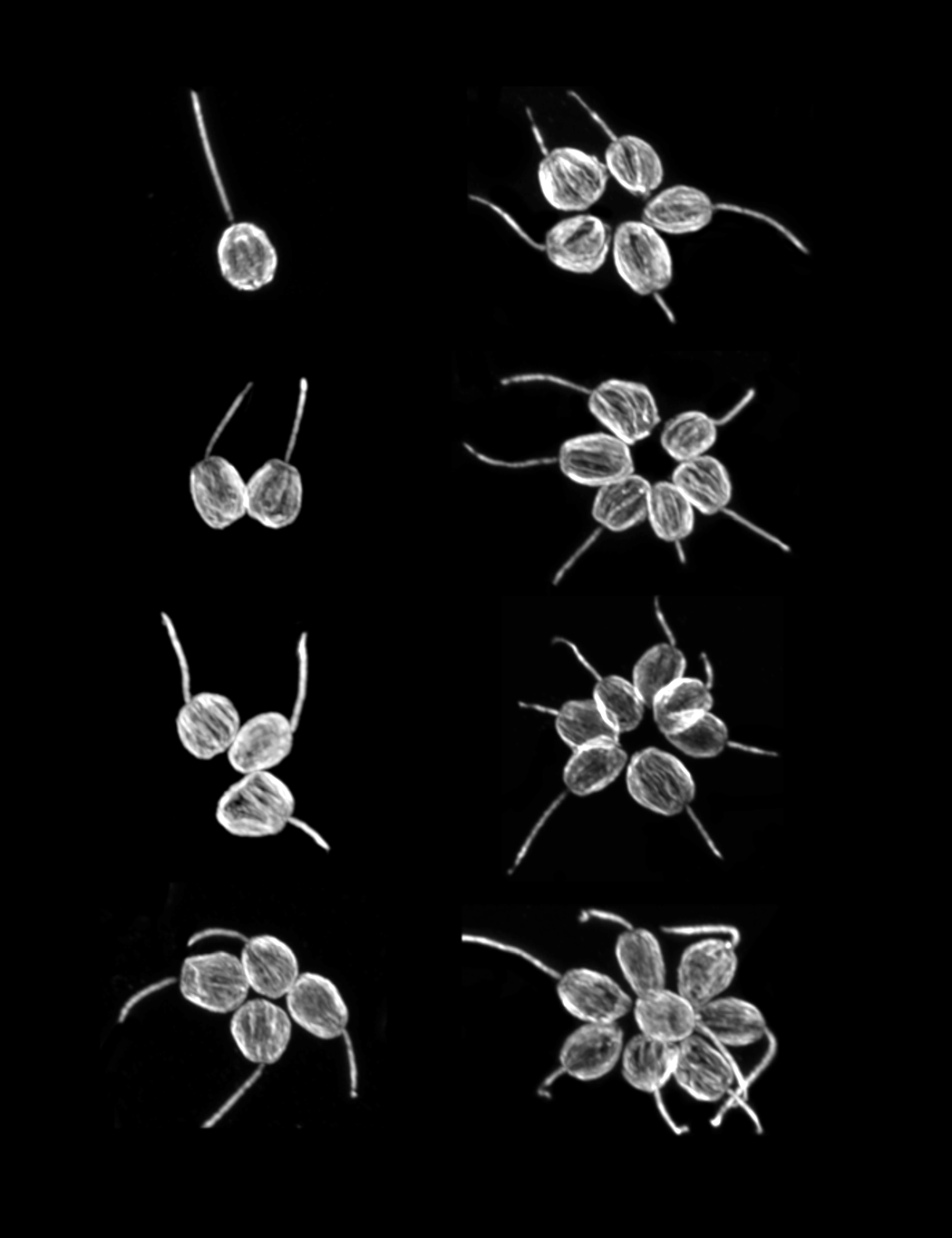
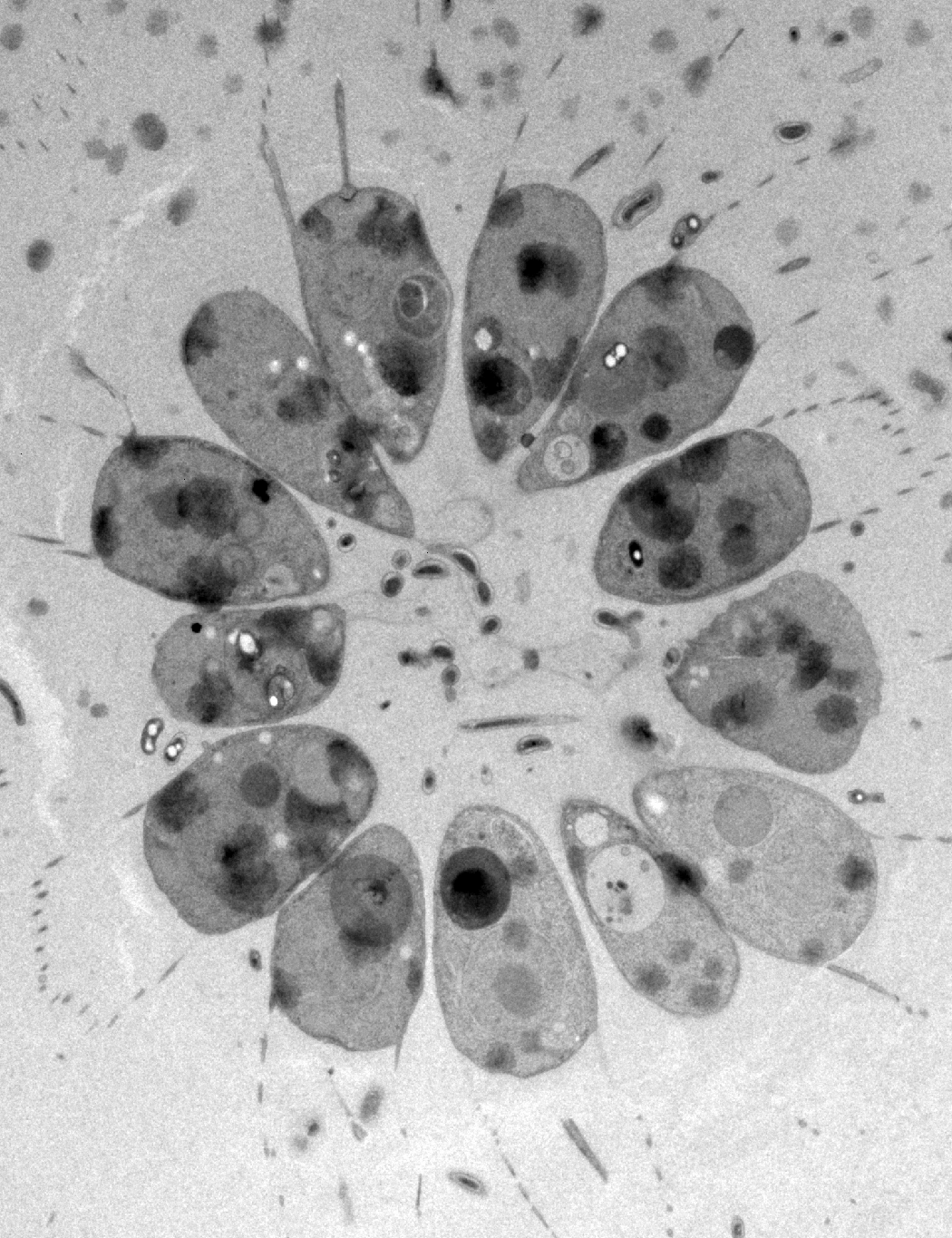
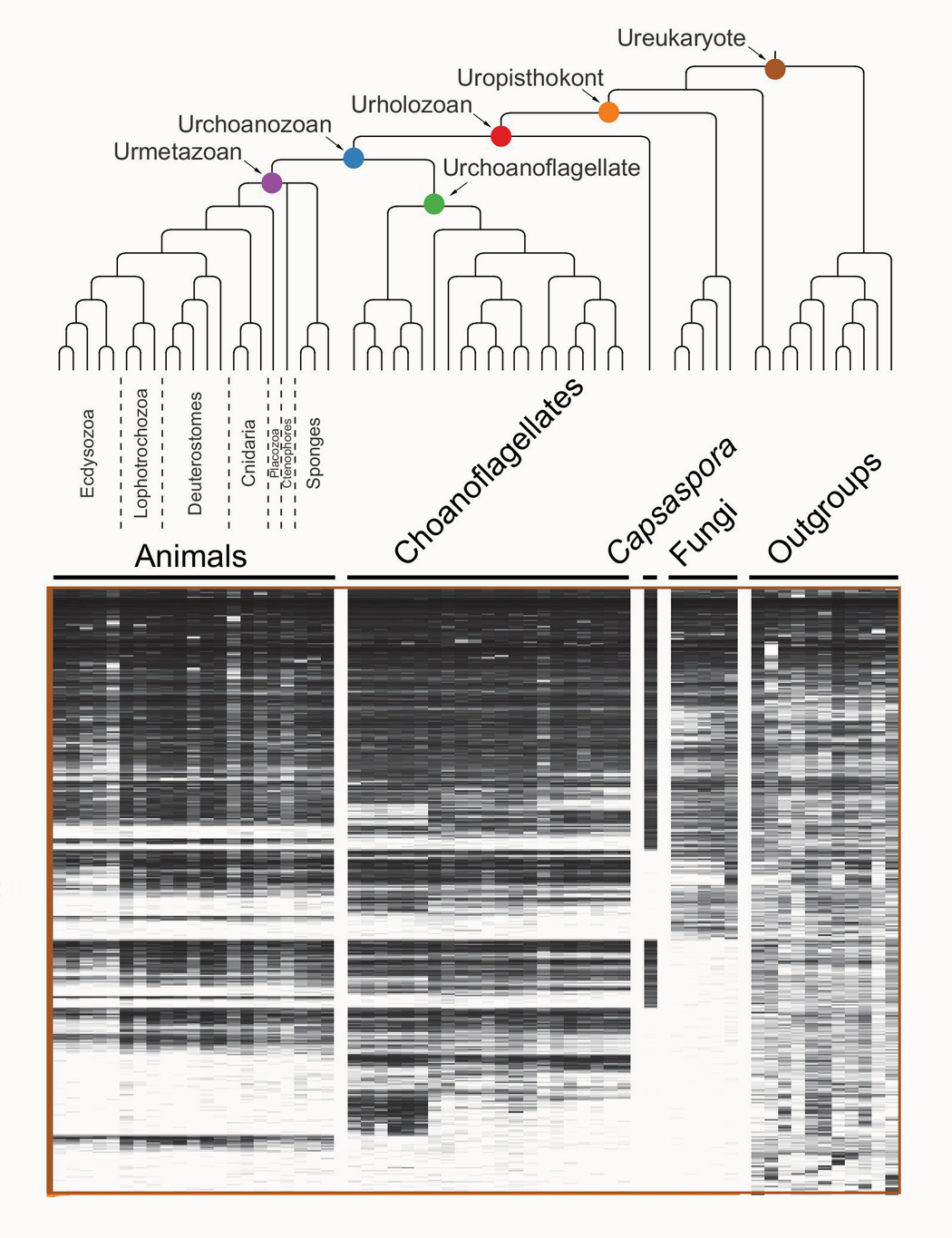
The origin of animals represents one of the pivotal transitions in life’s history, and one of its greatest unsolved mysteries. While the fossil record remains silent regarding the rise of multicellularity, the genetic and developmental foundations of animal origins may be deduced from shared elements among extant animals and their protozoan relatives, the choanoflagellates. To better understand the origin and evolution of animals, we are:
1. reconstructing the minimal genomic complexity of the unicellular progenitors of animals
2. elucidating the ancestral functions of genes required for animal development
3. characterizing choanoflagellate cell and developmental biology

Comments are closed.